Data set:
It is one of the challenges posed by Kaggle in the UCI Machine Learning repository
It is a dataset of breast cancer patients with malignant and benign tumors.
Logistic regression is used to predict whether a given patient will have a malignant or benign tumor based on the attributes in a given data set.
code:loading libraries
# performing linear algebra
import numpy as np
# data processing
import pandas as pd
# visualisation
import matplotlib.pyplot as pltCode: Load the dataset
data = pd.read_csv( "..\\breast-cancer-wisconsin-data\\data.csv" )
print (data.head)Output:
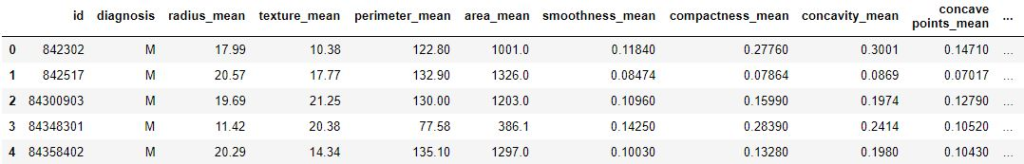
Code: Load the dataset
data.info()Output:
RangeIndex: 569 entries, 0 to 568
Data columns (total 33 columns):
id 569 non-null int64
diagnosis 569 non-null object
radius_mean 569 non-null float64
texture_mean 569 non-null float64
perimeter_mean 569 non-null float64
area_mean 569 non-null float64
smoothness_mean 569 non-null float64
compactness_mean 569 non-null float64
concavity_mean 569 non-null float64
concave points_mean 569 non-null float64
symmetry_mean 569 non-null float64
fractal_dimension_mean 569 non-null float64
radius_se 569 non-null float64
texture_se 569 non-null float64
perimeter_se 569 non-null float64
area_se 569 non-null float64
smoothness_se 569 non-null float64
compactness_se 569 non-null float64
concavity_se 569 non-null float64
concave points_se 569 non-null float64
symmetry_se 569 non-null float64
fractal_dimension_se 569 non-null float64
radius_worst 569 non-null float64
texture_worst 569 non-null float64
perimeter_worst 569 non-null float64
area_worst 569 non-null float64
smoothness_worst 569 non-null float64
compactness_worst 569 non-null float64
concavity_worst 569 non-null float64
concave points_worst 569 non-null float64
symmetry_worst 569 non-null float64
fractal_dimension_worst 569 non-null float64
Unnamed: 32 0 non-null float64
dtypes: float64(31), int64(1), object(1)
memory usage: 146.8+ KBCode: We removed the “id” and “Unnamed:32” columns because they have no role in the prediction
data.drop([ 'Unnamed: 32' , 'id' ], axis = 1 )
data.diagnosis = [ 1 if each = = "M" else 0 for each in data.diagnosis]Code: Input and output data
y = data.diagnosis.values
x_data = data.drop([ 'diagnosis' ], axis = 1 )code:normalizing
x = (x_data - np. min (x_data)) /(np. max (x_data) - np. min (x_data)).valuesCode: Split data for training and testing.
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size = 0.15 , random_state = 42 )
x_train = x_train.T
x_test = x_test.T
y_train = y_train.T
y_test = y_test.T
print ( "x train: " , x_train.shape)
print ( "x test: " , x_test.shape)
print ( "y train: " , y_train.shape)
print ( "y test: " , y_test.shape)Code: Weight and bias
def initialize_weights_and_bias(dimension):
w = np.full((dimension, 1 ), 0.01 )
b = 0.0
return w, bCode: Sigmoid function – calculates z-value.
# z = np.dot(w.T, x_train)+b
def sigmoid(z):
y_head = 1 /( 1 + np.exp( - z))
return y_headCode: Propagating from front to back
def forward_backward_propagation(w, b, x_train, y_train):
z = np.dot(w.T, x_train) + b
y_head = sigmoid(z)
loss = - y_train * np.log(y_head) - ( 1 - y_train) * np.log( 1 - y_head)
# x_train.shape[1] is for scaling
cost = (np. sum (loss)) /x_train.shape[ 1 ]
# backward propagation
derivative_weight = (np.dot(x_train, (
(y_head - y_train).T))) /x_train.shape[ 1 ]
derivative_bias = np. sum (
y_head - y_train) /x_train.shape[ 1 ]
gradients = { "derivative_weight" : derivative_weight, "derivative_bias" : derivative_bias}
return cost, gradientscode: Update parameters
def update(w, b, x_train, y_train, learning_rate, number_of_iterarion):
cost_list = []
cost_list2 = []
index = []
# updating(learning) parameters is number_of_iterarion times
for i in range (number_of_iterarion):
# make forward and backward propagation and find cost and gradients
cost, gradients = forward_backward_propagation(w, b, x_train, y_train)
cost_list.append(cost)
# lets update
w = w - learning_rate * gradients[ "derivative_weight" ]
b = b - learning_rate * gradients[ "derivative_bias" ]
if i % 10 = = 0 :
cost_list2.append(cost)
index.append(i)
print ( "Cost after iteration % i: % f" % (i, cost))
# update(learn) parameters weights and bias
parameters = { "weight" : w, "bias" : b}
plt.plot(index, cost_list2)
plt.xticks(index, rotation = 'vertical' )
plt.xlabel( "Number of Iterarion" )
plt.ylabel( "Cost" )
plt.show()
return parameters, gradients, cost_listCode: Prediction
def predict(w, b, x_test):
# x_test is a input for forward propagation
z = sigmoid(np.dot(w.T, x_test) + b)
Y_prediction = np.zeros(( 1 , x_test.shape[ 1 ]))
# if z is bigger than 0.5, our prediction is sign one (y_head = 1), # if z is smaller than 0.5, our prediction is sign zero (y_head = 0), for i in range (z.shape[ 1 ]):
if z[ 0 , i]<= 0.5 :
Y_prediction[ 0 , i] = 0
else :
Y_prediction[ 0 , i] = 1
return Y_predictionCode: Logistic Regression
def logistic_regression(x_train, y_train, x_test, y_test, learning_rate, num_iterations):
dimension = x_train.shape[ 0 ]
w, b = initialize_weights_and_bias(dimension)
parameters, gradients, cost_list = update(
w, b, x_train, y_train, learning_rate, num_iterations)
y_prediction_test = predict(
parameters[ "weight" ], parameters[ "bias" ], x_test)
y_prediction_train = predict(
arameters[ "weight" ], parameters[ "bias" ], x_train)
# train /test Errors
print ( "train accuracy: {} %" . format (
100 - np.mean(np. abs (y_prediction_train - y_train)) * 100 ))
print ( "test accuracy: {} %" . format (
100 - np.mean(np. abs (y_prediction_test - y_test)) * 100 ))
logistic_regression(x_train, y_train, x_test, y_test, learning_rate = 1 , num_iterations = 100 )Output:
Cost after iteration 0: 0.692836
Cost after iteration 10: 0.498576
Cost after iteration 20: 0.404996
Cost after iteration 30: 0.350059
Cost after iteration 40: 0.313747
Cost after iteration 50: 0.287767
Cost after iteration 60: 0.268114
Cost after iteration 70: 0.252627
Cost after iteration 80: 0.240036
Cost after iteration 90: 0.229543
Cost after iteration 100: 0.220624
Cost after iteration 110: 0.212920
Cost after iteration 120: 0.206175
Cost after iteration 130: 0.200201
Cost after iteration 140: 0.194860
Output:
train accuracy: 95.23809523809524 %
test accuracy: 94.18604651162791 %code:using linear_model. LogisticRegression test results
from sklearn import linear_model
logreg = linear_model.LogisticRegression(random_state = 42 , max_iter = 150 )
print ( "test accuracy: {} " . format (
logreg.fit(x_train.T, y_train.T).score(x_test.T, y_test.T)))
print ( "train accuracy: {} " . format (
logreg.fit(x_train.T, y_train.T).score(x_train.T, y_train.T)))Output:
test accuracy: 0.9651162790697675
train accuracy: 0.9668737060041408First, your interview preparation enhances your data structure concepts with the Python DS course.